Abstract
Introduction: Measurable residual disease (MRD) detection is associated with relapse. Leukemias with KMT2A rearrangement (KMT2Ar) have an adverse prognosis with more than 80 possible fusion partner genes (FPG) identified. With the advent of effective targeted therapies for KMT2Ar leukemia such as menin inhibitors, there is an unmet need for sensitive assays to detect these various fusions, not only to guide and monitor therapy, but also to predict and intercept relapse. Multicolor flow cytometry (MFC) is the only readily available test for MRD in KMT2Ar acute myeloid leukemia (AML), not requiring prior knowledge of exact fusions and breakpoints for design of patient-specific assays. Standard next-generation sequencing (NGS) methods cannot accurately detect genetic alterations at low frequencies because of inherent technical errors and the need for high sequencing depth. Blocker Displacement Amplification (BDA) technology enables selective detection of such rare alterations with NGS (Wu LR, Nat Biomed Eng 2017). Here, we demonstrate the effective detection of KMT2Ar and its various FPG with a novel NGS assay leveraging BDA.
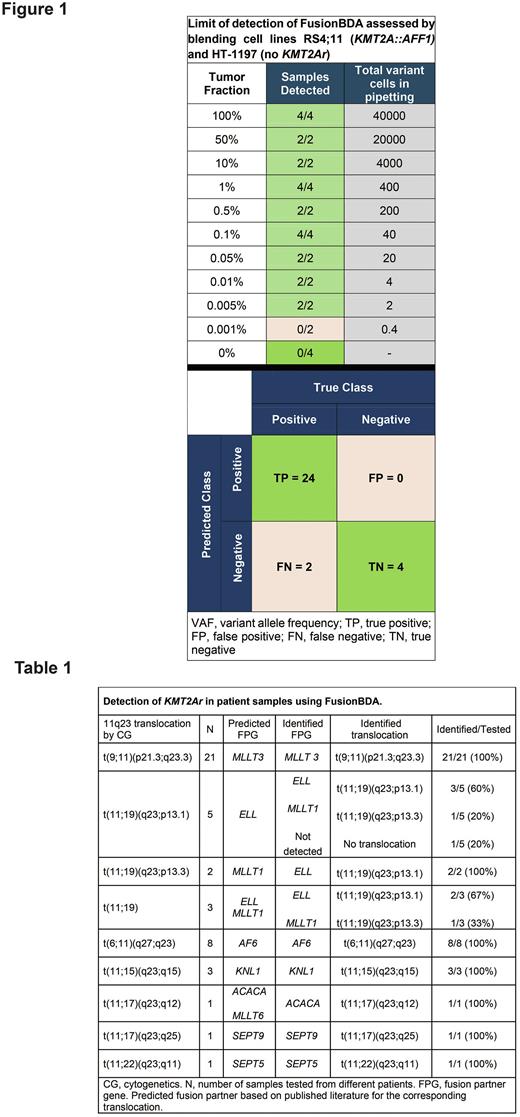
Methods: The NuProbe RNA fusion proprietary technology uses BDA (FusionBDA) where primers flank any known exon junction of cDNA corresponding to transcripts of the gene of interest, while non-extensible oligonucleotides (blockers) span the wildtype (WT) exon junction, therefore suppressing amplification of WT while differentially amplifying fusion junctions. As a result, final libraries are enriched with reads mapping to fusion genes. This allows de novo calls and minimizes read requirements. We designed a custom panel targeting 35 exons of KMT2A (forward and reverse) and developed a pipeline using 3 previously established fusion detection methods (Arriba, STAR-Fusion, FusionCatcher). We validated this assay in a KMT2Ar cell line. Limit-of-detection (LoD) was assessed by blending RS4;11 (leukemia with KMT2A::AFF1) with HT-1197 cells (bladder cancer, no KMT2Ar), to attain contrived samples with 0.001% to 100% tumor fractions (400ng input). Limit-of-input (LoI) was assessed using cDNA with 1- 400ng RNA from 100% and 0.1% tumor fraction samples. In a blinded analysis, we tested 50 bone marrow samples from 35 patients with KMT2Ar AML, chosen deliberately with various 11q23 translocations including rare ones.
Results: Following LoD experiment, KMT2Ar was detected accurately down to 0.005% tumor fraction (Figure 1). Subsequently, LoI assay revealed the ability to detect KMT2Ar in 100% VAF tumor fraction samples using as low as 1ng of input RNA and using 400ng in 1% VAF samples.
Among samples with morphologic evidence of leukemia, FusionBDA detected various KMT2Ar in 44/45 samples (97.8%) except 1 sample with a t(11;19)(q23;p13.1) (Table 1). This sample had a suboptimal amount and quality of cDNA loaded. We evaluated concordance of predicted FPG based on bands by cytogenetics (CG) and the published literature on KMT2A recombinome. We found that FusionBDA identified the same predicted FPG (same band) in 41/45 samples (91%). In samples with various t(11;19) translocations, different FPG were identified all located on 11p13 within a few bands from ones by CG, likely representing the accurate FPG given the low resolution of CG and the associated error in determining arm band accurately.
Finally, we explored the potential of this assay for MRD detection by examining 5 samples at morphologic remission following treatment. All samples where fluorescence in situ hybridization (FISH) detected a KMT2A fusion had FusionBDA+ (31/31 samples including 1 in morphologic remission). 1 sample with no detectable KMT2A fusion by FISH and MRDneg by MFC, had detectable fusion by FusionBDA, indicating possibly an improved sensitivity. 3 samples with MRD+ by MFC also had detectable KMT2Ar by FusionBDA (lowest MRD+ level by flow was 0.5%). 1 sample was MRD+ by MFC and had a detectable KMT2Ar by FusionBDA in 2/3 callers (detectable by FusionCatcher and StarFusion not Arriba), therefore not meeting our stringent criteria though likely concordant with MRD by MFC. Further analysis in the MRD setting is planned.
Conclusion: FusionBDA is a novel, sensitive, and specific assay for KMT2Ar acute leukemia. This assay is able to agnostically detect KMT2Ar with various fusions at a predicted sensitivity of at least 0.005%, therefore allowing both MRD detection and target identification.
Disclosures
Issa:Celgene, Kura Oncology, Syndax, Merck, Cullinan and Novartis: Research Funding; Novartis, Kura Oncology, Nuprobe: Consultancy. Roberts:NuProbe: Current Employment. Li:NuProbe: Current Employment. Weller:NuProbe: Current Employment. Alam:NuProbe: Current Employment. Gonzalez:NuProbe: Current Employment. Maupin:NuProbe: Current Employment. Nguyen:NuProbe: Current Employment. Guzman:NuProbe: Current Employment. Nguyen:NuProbe: Current Employment. Casas Lumbreras:NuProbe: Ended employment in the past 24 months. Thirunavukarasu:NuProbe: Current Employment. Pinto:NuProbe: Current Employment. Zhang:NuProbe: Consultancy, Current Employment, Current equity holder in private company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal